In the field of proteomics research, the quest for a platform that combines high-throughput capability, specificity, and sensitivity is ongoing. Olink Explore has emerged as a promising innovation in the fields of proteomics and biomarker discovery, offering a sophisticated solution to scientists and researchers dedicated to advancing our understanding of proteins and their roles in disease and drug development. In this blog post we delve into the Olink Explore platform, its underlying technology and benefits, and how it serves as a pivotal tool in advancing proteomics research.
At the heart of the Olink Explore platform lies the Proximity Extension Assay (PEA) technology. PEA technology is a sophisticated biochemical process that uniquely combines the specificity of antibodies with the sensitivity and amplification capabilities of PCR (Polymerase Chain Reaction). The core mechanism of PEA involves two proximity probes, each consisting of an antibody linked to a unique DNA oligonucleotide [1]. When these probes bind to their target protein, they are brought into close proximity. This proximity allows the DNA oligonucleotides to hybridize, forming a double-stranded DNA molecule that can be extended by a DNA polymerase. Sample multiplexing is enabled by adding unique sample indices during a subsequent PCR step. This process results in the generation of a unique DNA sequence that is directly quantifiable using real-time PCR or Next-Generation Sequencing (NGS), translating protein presence into a measurable DNA signal. Using this method the NGS counts are proportional to the protein’s concentration in the sample, enabling its relative quantification (Figure 1).
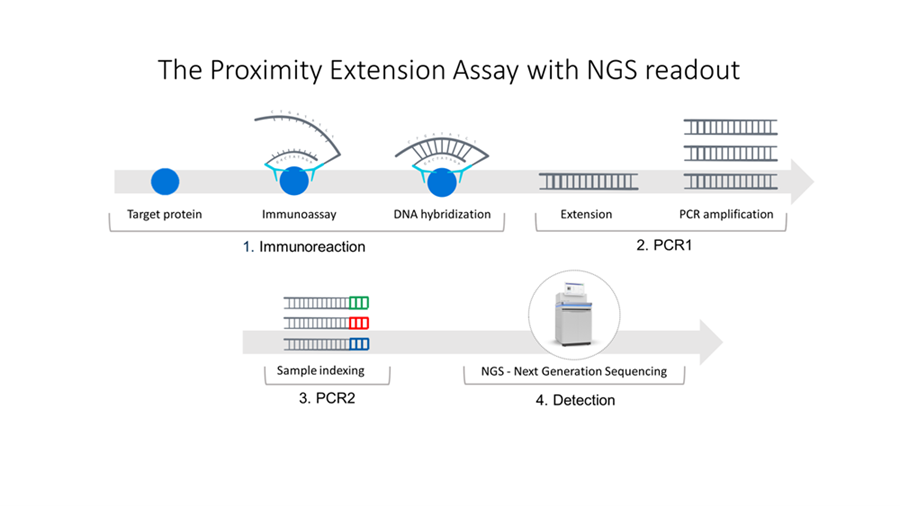
One of the distinguishing features of PEA technology is its requirement for dual binding, significantly reducing the risk of nonspecific signals and thus enhancing the assay’s specificity. This dual-recognition strategy ensures that only the correct target proteins are detected, minimizing the chances of false positives. Moreover, the amplification step allows for the detection of low abundance proteins, addressing a common challenge in proteomics research-sensitivity.
Over the years, PEA technology has evolved and been refined, benefiting from advancements in antibody engineering, DNA synthesis, and amplification techniques. This evolution has expanded its applications, making it a versatile tool for biomarker discovery, disease research, and drug development. The integration of PEA with NGS readouts, as seen in the Olink Explore platform, represents a significant leap forward, enabling unprecedented multiplexing capabilities and depth of analysis.
More specifically, Olink Explore can accelerate proteomics research by offering:
- Broad Dynamic Range: A 10-log dynamic range that facilitates the detection of both high and low abundant proteins.
- High Throughput: Capable of generating millions of data points per run, supporting large-scale studies.
- Improvements in Specificity and Sensitivity: The requirement of three binding events (antibody pair binding and oligo hybridization) coupled with signal amplification ensures high fidelity in protein detection.
- High Multiplex Capability: With the ability to measure up to 5700 targets, researchers can explore complex protein networks and pathways in a single run.
- Minimal Sample Consumption: Requires only 6-15ul of sample, preserving valuable research material.
- Versatile Sample Compatibility: Supports a wide range of sample types, from human plasma and serum to tissue lysates and beyond, accommodating diverse research contexts.
A testament to the transformative impact of Olink Explore in the field of biomedical research is its significant contribution to the UK Biobank study. The UK Biobank Pharma Proteomics Project (UKB-PPP) represents a milestone in proteomics research, showcasing the potential of Olink Explore in uncovering the intricate relationship of proteins and genetics. In a large-scale collaborative effort, 13 biopharmaceutical companies utilized the Olink Explore platform to analyze up to 3,000 proteins across all major biological pathways in over 54,000 participant samples from the UK Biobank. This comprehensive study, detailed in a pivotal Nature article, not only underscores the platform’s high-throughput capability but also its crucial role in advancing our understanding of proteome-genome interactions [2].
In terms of protein biomarkers, Olink’s assay library is carefully curated, covering major biological pathways and indications relevant to cardiometabolics, oncology, inflammation, and neurology. This comprehensive resource includes exploratory proteins, known drug targets, diagnostic biomarkers, and disease-related proteins, offering a solid foundation for targeted and exploratory research.
Olink Explore places a high value on quality and transparency. Each assay undergoes rigorous quality control, with validation data readily accessible. Researchers have open access to validation information, including calibration curves, limits of detection (LOD), and precision metrics for each assay ensuring confidence in their experimental data. Additionally, the scalability of Olink Explore NGS platform is underscored by its high correlation with the qPCR-based Olink platform (Olink Target), boasting a median coefficient of 0.95. This seamless integration facilitates the expansion of studies from targeted to broad analyses without sacrificing data integrity.
Olink Explore by Protavio: Pioneering the Future of Proteomics
At Protavio, our mission-proteomics for better health-drives our commitment to advancing the frontiers of protein research. Understanding the critical role of proteomics in unraveling the complexities of health and disease, we continuously seek to be at the forefront of technological advancement. It is with this vision that we have recognized the innovation and potential of the Olink Explore platform, a decision that significantly enhances our capabilities in novel protein biomarker discovery. Leveraging our deep proteomics expertise in the Luminex platform and our deep understanding of diagnostic product development, our alignment with Olink Explore allows us to offer a broad spectrum of services tailored to the diverse needs of the scientific community. From biomarker discovery to the development and manufacturing of diagnostic assays, our partnerships enable us to provide comprehensive solutions that drive forward the pursuit of better health through proteomics.
Protavio is a certified service provider and offers the Olink Explore platform in three tailored solutions, each designed to meet specific research requirements and sample throughputs:
- Explore Individual 384x Target Kits: Catering to focused studies, these kits allow for the analysis of 384 proteins across a wide array of indications, offering precision and depth in targeted research.
- Explore 3k: A robust solution comprising eight 384x target kits, the Explore 3k is the choice for comprehensive studies, facilitating extensive protein analysis with unparalleled breadth.
- Explore HT: For researchers pushing the boundaries of proteomics, the Explore HT provides the ultimate high-throughput option. Capable of measuring up to 5700 targets across a minimum of 176 samples, it is ideally suited for large-scale investigations seeking to uncover the proteome’s vast potential.
Having undergone rigorous training and successfully passed all requisite tests, Protavio is not just a provider but a trusted partner in the Olink Explore ecosystem. Our certified service offering underscores our commitment to excellence and our dedication to empowering researchers with the tools and insights needed to advance the field of proteomics.
References
- Lundberg, Martin, et al. “Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood.” Nucleic acids research 39.15 (2011): e102-e102.
- Sun, Benjamin B., et al. “Plasma proteomic associations with genetics and health in the UK Biobank.” Nature 622.7982 (2023): 329-338.
